Playing with Variational Autoencoders
[1]:
import os
import numpy as np
import pandas as pd
import tensorflow as tf
import tensorflow_probability as tfp
from sklearn.decomposition import PCA
import seaborn as sns
import matplotlib.pyplot as plt
from tqdm.auto import tqdm, trange
[2]:
tfd = tfp.distributions
sns.set_context('talk')
Glossary
epoch: one cycle through training data (made up of many steps)
step: one gradient update per batch of data
learning rate: how fast to follow gradient (in gradient descent)
neural network: find function \(y = f(x)\) with features \(x\) and values/labels \(y\) (regression/classification)
autoencoder:
find function mapping \(x\) to itself: \(z = f(h_e(x))\), \(\hat{x} = f(h_d(z))\)
undercomplete: \(dim(latent\ space) < dim(input\ space)\) (vs overcomplete)
problem of capacity:
if encoder is too powerful, one dimensional latent space could encode all training samples -> autoencoder fails to learn anything useful
regularized: sophisticated loss-function (sparsity, robustness, …)
variational: enforce distribution for latent space
Introduction
Basic operations
[3]:
a = tf.constant(2.0)
b = tf.constant(3.0)
print(tf.add(a, b))
print(tf.reduce_mean([a, b]))
tf.Tensor(5.0, shape=(), dtype=float32)
tf.Tensor(2.5, shape=(), dtype=float32)
[4]:
m1 = tf.constant([[1.0, 2.0], [3.0, 4.0]])
m2 = tf.constant([[5.0, 6.0], [7.0, 8.0]])
tf.matmul(m1, m2)
[4]:
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[19., 22.],
[43., 50.]], dtype=float32)>
Variational Autoencoder
Theory
Given data \(X\), latent variable \(z\), probability distribution of data \(P(X)\), p.d. of latent variable \(P(z)\), p.d. of generated data given latent variable \(P(X|z)\)
Goal is to model data, i.e. find \(P(X) = \int P(X|z) P(z)\)
Idea of VAE:
infer \(P(z)\) from \(P(z|X)\) using variational inference (inference as optimization problem)
model \(P(z|X)\) using \(Q(z|X)\), \(Q\) is typically Gaussian
components:
encoder: \(Q(z|X)\)
decoder: \(P(X|z)\)
Measure difference between two distributions: Kullback–Leibler divergence
Objective function:
Design model
[5]:
class Encoder(tf.keras.layers.Layer):
"""Convert input to low-dimensional representation."""
def __init__(self, latent_dim, original_dim):
super(Encoder, self).__init__()
self.network = tf.keras.Sequential(
[
tf.keras.layers.InputLayer(input_shape=(*original_dim, 1)),
tf.keras.layers.Conv2D(
filters=32, kernel_size=3, strides=(2, 2), activation=tf.nn.relu
),
tf.keras.layers.Conv2D(
filters=64, kernel_size=3, strides=(2, 2), activation=tf.nn.relu
),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(units=2 * latent_dim),
]
)
def call(self, x):
return self.network(x)
class Decoder(tf.keras.layers.Layer):
"""Reconstruct input from low-dimensional representation."""
def __init__(self, latent_dim, original_dim):
super(Decoder, self).__init__()
self.network = tf.keras.Sequential(
[
tf.keras.layers.InputLayer(input_shape=(latent_dim,)),
tf.keras.layers.Dense(units=7 * 7 * 32, activation=tf.nn.relu),
tf.keras.layers.Reshape(target_shape=(7, 7, 32)),
tf.keras.layers.Conv2DTranspose(
filters=64,
kernel_size=3,
strides=(2, 2),
padding='SAME',
activation=tf.nn.relu,
),
tf.keras.layers.Conv2DTranspose(
filters=32,
kernel_size=3,
strides=(2, 2),
padding='SAME',
activation=tf.nn.relu,
),
tf.keras.layers.Conv2DTranspose(
filters=1, kernel_size=3, strides=(1, 1), padding='SAME'
),
]
)
def call(self, z):
return self.network(z)
class Autoencoder(tf.keras.Model):
"""Connect all components to single model."""
def __init__(self, latent_dim, original_dim):
"""Initialize everything."""
super(Autoencoder, self).__init__()
self.latent_dim = latent_dim
# setup architecture
self._encoder = Encoder(latent_dim=self.latent_dim, original_dim=original_dim)
self._decoder = Decoder(latent_dim=self.latent_dim, original_dim=original_dim)
# helpful stuff
self.train_loss = tf.keras.metrics.Mean(name='train_loss')
def encode(self, x):
"""Input -> latent distribution."""
mean, logvar = tf.split(self._encoder(x), num_or_size_splits=2, axis=1)
return mean, logvar
def _reparameterize(self, mean, logvar):
"""Trick which is needed for backpropagation."""
eps = tf.random.normal(shape=mean.shape)
return eps * tf.exp(logvar * 0.5) + mean
def decode(self, z):
"""Latent -> input space."""
return self._decoder(z)
def call(self, x):
"""Generate distribution from input and reconstruct using it."""
mu, _ = tf.split(self._encoder(x), num_or_size_splits=2, axis=1)
# why do we only pass mu?
reconstructed = self.decode(mu)
return reconstructed
def log_normal_pdf(self, sample, mean, logvar, raxis=1):
log2pi = tf.math.log(2.0 * np.pi)
return tf.reduce_sum(
-0.5 * ((sample - mean) ** 2.0 * tf.exp(-logvar) + logvar + log2pi),
axis=raxis,
)
def compute_loss(self, x):
"""Compute loss.
Hmmmmm
"""
mean, logvar = self.encode(x)
z = self._reparameterize(mean, logvar)
x_logit = self.decode(z)
cross_ent = tf.nn.sigmoid_cross_entropy_with_logits(logits=x_logit, labels=x)
logpx_z = -tf.reduce_sum(cross_ent, axis=[1, 2, 3])
logpz = self.log_normal_pdf(z, 0.0, 0.0)
logqz_x = self.log_normal_pdf(z, mean, logvar)
return -tf.reduce_mean(logpx_z + logpz - logqz_x)
def compute_gradients(self, x):
with tf.GradientTape() as tape:
loss = self.compute_loss(x)
self.train_loss(loss)
return tape.gradient(loss, self.trainable_variables)
@tf.function
def train(self, x, optimizer):
gradients = self.compute_gradients(x)
optimizer.apply_gradients(zip(gradients, self.trainable_variables))
def sample(self, eps=None):
if eps is None:
eps = tf.random.normal(shape=(100, self.latent_dim))
return self.decode(eps)
def save(self, fname):
"""Save model.
https://www.tensorflow.org/alpha/guide/keras/saving_and_serializing#saving_subclassed_models
"""
os.makedirs(os.path.dirname(fname), exist_ok=True)
self.save_weights(fname, save_format='tf')
@classmethod
def load_from_file(cls, fname):
model = cls(latent_dim, original_dim) # TODO: load parameters from file
# train model briefly to infer architecture
# TODO
model.load_weights(fname)
return model
Run model
Parameters
[6]:
np.random.seed(1)
tf.random.set_seed(1)
[7]:
batch_size = 128
epochs = 15
learning_rate = 1e-3
latent_dim = 10
original_dim = (28, 28)
Load data
[8]:
(training_features, training_labels), (
test_features,
_,
) = tf.keras.datasets.mnist.load_data()
# normalize to range [0, 1]
training_features = training_features / np.max(training_features)
# flatten 2D images into 1D
training_features = training_features.reshape(
training_features.shape[0], *original_dim, 1
).astype(np.float32)
# prepare dataset
training_dataset = tf.data.Dataset.from_tensor_slices(training_features).batch(
batch_size
)
training_dataset
[8]:
<BatchDataset shapes: (None, 28, 28, 1), types: tf.float32>
Train model
[9]:
autoencoder = None
[10]:
opt = tf.optimizers.Adam(learning_rate=learning_rate)
[11]:
# TODO: improve loss storage
fname = 'models/autoencoder'
if os.path.exists(f'{fname}.index'):
autoencoder = Autoencoder.load_from_file(fname)
df_loss = pd.read_csv('models/autoencoder_loss.csv')
else:
autoencoder = Autoencoder(latent_dim=latent_dim, original_dim=original_dim)
# train
loss_list = []
for epoch in trange(epochs, desc='Epochs'):
for step, batch_features in enumerate(training_dataset):
autoencoder.train(batch_features, opt)
loss_list.append(
{'epoch': epoch, 'loss': autoencoder.train_loss.result().numpy()}
)
df_loss = pd.DataFrame(loss_list)
# save
autoencoder.save(fname)
df_loss.to_csv('models/autoencoder_loss.csv', index=False)
WARNING:tensorflow:Inconsistent references when loading the checkpoint into this object graph. Either the Trackable object references in the Python program have changed in an incompatible way, or the checkpoint was generated in an incompatible program.
Two checkpoint references resolved to different objects (<tensorflow.python.keras.layers.convolutional.Conv2D object at 0x163057d30> and <tensorflow.python.keras.layers.convolutional.Conv2D object at 0x163057ee0>).
WARNING:tensorflow:Inconsistent references when loading the checkpoint into this object graph. Either the Trackable object references in the Python program have changed in an incompatible way, or the checkpoint was generated in an incompatible program.
Two checkpoint references resolved to different objects (<tensorflow.python.keras.layers.convolutional.Conv2D object at 0x163057ee0> and <tensorflow.python.keras.layers.core.Flatten object at 0x163057ac0>).
WARNING:tensorflow:Inconsistent references when loading the checkpoint into this object graph. Either the Trackable object references in the Python program have changed in an incompatible way, or the checkpoint was generated in an incompatible program.
Two checkpoint references resolved to different objects (<tensorflow.python.keras.layers.core.Dense object at 0x16312d730> and <tensorflow.python.keras.layers.core.Reshape object at 0x16312ddc0>).
WARNING:tensorflow:Inconsistent references when loading the checkpoint into this object graph. Either the Trackable object references in the Python program have changed in an incompatible way, or the checkpoint was generated in an incompatible program.
Two checkpoint references resolved to different objects (<tensorflow.python.keras.layers.convolutional.Conv2DTranspose object at 0x16312df10> and <tensorflow.python.keras.layers.convolutional.Conv2DTranspose object at 0x16314a4c0>).
WARNING:tensorflow:Inconsistent references when loading the checkpoint into this object graph. Either the Trackable object references in the Python program have changed in an incompatible way, or the checkpoint was generated in an incompatible program.
Two checkpoint references resolved to different objects (<tensorflow.python.keras.layers.convolutional.Conv2DTranspose object at 0x16314a4c0> and <tensorflow.python.keras.layers.convolutional.Conv2DTranspose object at 0x16314aa00>).
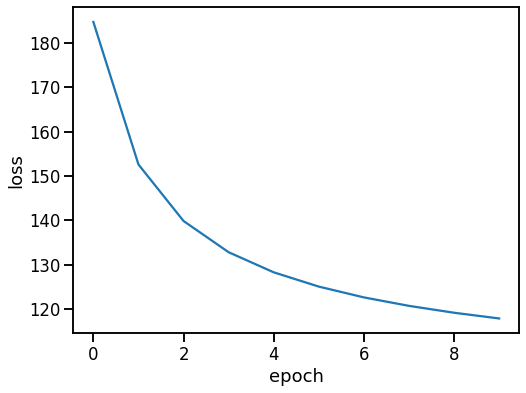
Analysis
Loss development
[12]:
plt.figure(figsize=(8, 6))
sns.lineplot(x='epoch', y='loss', data=df_loss)
[12]:
<AxesSubplot:xlabel='epoch', ylabel='loss'>
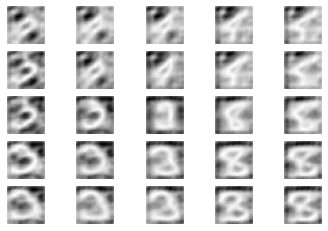
Generate new samples
[13]:
# inp = tf.random.normal(shape=(4, latent_dim), mean=10)
inp = np.asarray([[0.0] * latent_dim])
inp[0][4] = -2
[14]:
N = 5
fig, ax_arr = plt.subplots(N, N)
for i, x in enumerate(np.linspace(-10, 10, N)):
for j, y in enumerate(np.linspace(-10, 10, N)):
ax = ax_arr[i, j]
inp[0][:2] = np.random.normal([x, y])
img = autoencoder.sample(inp).numpy()
ax.imshow(img[0, :, :, 0], cmap='gray')
ax.axis('off')
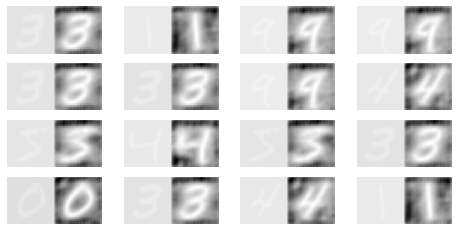
Check specific examples
[15]:
features_sub = training_features[:10]
features_sub.shape
[15]:
(10, 28, 28, 1)
[16]:
original = tf.reshape(features_sub, (features_sub.shape[0], 28, 28))
reconstructed = tf.reshape(
autoencoder(tf.constant(features_sub)), (features_sub.shape[0], 28, 28)
)
[17]:
N = 16
fig, ax_list = plt.subplots(nrows=4, ncols=4, figsize=(8, 4))
for i, ax in enumerate(ax_list.ravel()):
idx = np.random.randint(0, original.shape[0])
img_orig = original.numpy()[idx]
img_recon = reconstructed.numpy()[idx]
img_concat = np.concatenate([img_orig, img_recon], axis=1)
ax.imshow(img_concat, cmap='gray')
ax.axis('off')
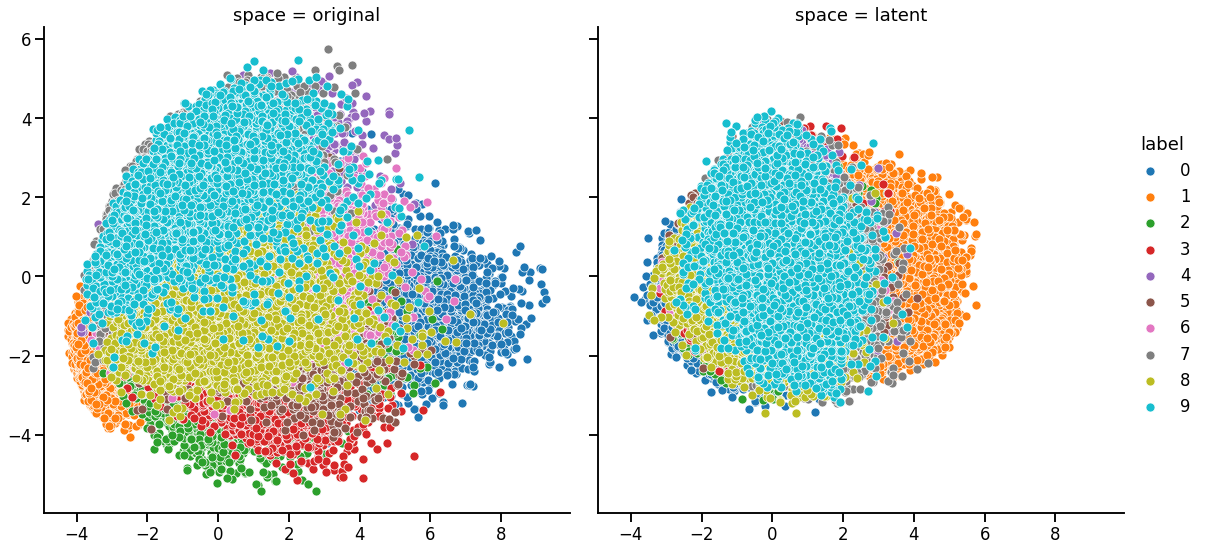
PCA
[18]:
def do_PCA(X, ndim=2):
pca = PCA(n_components=ndim)
pca.fit(X)
X_trans = pca.transform(X)
print(pca.explained_variance_ratio_)
return pd.DataFrame(X_trans, columns=[f'PC_{i}' for i in range(ndim)])
[19]:
latent_features = autoencoder.encode(training_features)
latent_features_1d = np.concatenate(latent_features, axis=1)
df_latent = do_PCA(latent_features_1d)
df_latent['label'] = training_labels
df_latent['space'] = 'latent'
[0.20341446 0.12413041]
[20]:
data_train_1d = training_features.reshape(
training_features.shape[0], training_features.shape[1] * training_features.shape[2]
)
df_original = do_PCA(data_train_1d)
df_original['label'] = training_labels
df_original['space'] = 'original'
[0.09704707 0.07095956]
[21]:
df_pca = pd.concat([df_original, df_latent])
[22]:
g = sns.FacetGrid(df_pca, col='space', hue='label', height=8, legend_out=True)
g.map_dataframe(sns.scatterplot, x='PC_0', y='PC_1', rasterized=True)
g.add_legend()
[22]:
<seaborn.axisgrid.FacetGrid at 0x163c546a0>