Visualizing Eurostat data
[2]:
import numpy as np
import pandas as pd
import geopandas as gpd
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.pyplot import cm
import geoplot as gplt
import geoplot.crs as gcrs
from tqdm.auto import tqdm
from rwrap import eurostat
[3]:
sns.set_context('talk')
Retrieving geographical information
The NUTS classification subdivides each member state into regions at three different levels, covering NUTS 1, 2 and 3 from larger to smaller areas (https://ec.europa.eu/eurostat/web/nuts/background). Eurostat provides this geographical data in various formats (https://ec.europa.eu/eurostat/web/gisco/geodata/reference-data/administrative-units-statistical-units, looks for “NUTS”).
The filename of each dataset follows a specific format: <theme>_<spatialtype>_<resolution>_<year>_<projection>_<subset>.<extension> (check https://ec.europa.eu/eurostat/cache/GISCO/distribution/v2/nuts/nuts-2016-files.html for more information).
To make things easier, we use the eurostat package to retrieve this data automatically.
[5]:
df_geo = eurostat.get_eurostat_geospatial(
output_class='sf', resolution=60, nuts_level=2, year=2016
)
df_geo.head()
[5]:
| id | CNTR_CODE | NUTS_NAME | LEVL_CODE | FID | NUTS_ID | geometry | geo | |
|---|---|---|---|---|---|---|---|---|
| 0 | DE24 | DE | Oberfranken | 2 | DE24 | DE24 | POLYGON ((11.48157 50.43162, 11.48290 50.40161... | DE24 |
| 1 | NO03 | NO | Sør-Østlandet | 2 | NO03 | NO03 | POLYGON ((10.60072 60.13161, 10.57604 60.11572... | NO03 |
| 2 | HU11 | HU | Budapest | 2 | HU11 | HU11 | POLYGON ((18.93274 47.57117, 19.02713 47.58717... | HU11 |
| 3 | HU12 | HU | Pest | 2 | HU12 | HU12 | POLYGON ((18.96591 47.02896, 18.89943 47.07825... | HU12 |
| 4 | HU21 | HU | Közép-Dunántúl | 2 | HU21 | HU21 | POLYGON ((18.68843 47.57707, 18.70448 47.52134... | HU21 |
Example
To get a feeling for the data, we can plot the geographical composition of an arbitrary country.
[7]:
ax = gplt.polyplot(df_geo[df_geo['CNTR_CODE'] == 'DE'])
ax.set_aspect(1.4)
Retrieving statistical data
Eurostat is a great source for data related to Europe. An overview of the main tables can be found on https://ec.europa.eu/eurostat/web/regions/data/main-tables.
Select datasets
[8]:
eurostat.search_eurostat('age').head()
[8]:
| title | code | type | last update of data | last table structure change | data start | data end | values | |
|---|---|---|---|---|---|---|---|---|
| 1 | Manure storage facilities by NUTS 3 regions | aei_fm_ms | dataset | 05.02.2019 | 27.02.2020 | 2000 | 2010 | None |
| 2 | Population on 1 January by age, sex and NUTS 2... | demo_r_d2jan | dataset | 13.10.2020 | 16.06.2020 | 1990 | 2019 | None |
| 3 | Population on 1 January by age group, sex and ... | demo_r_pjangroup | dataset | 18.06.2020 | 14.05.2020 | 1990 | 2019 | None |
| 4 | Population on 1 January by age group, sex and ... | demo_r_pjangrp3 | dataset | 13.10.2020 | 14.05.2020 | 2014 | 2019 | None |
| 5 | Population on 1 January by broad age group, se... | demo_r_pjanaggr3 | dataset | 16.06.2020 | 16.06.2020 | 1990 | 2019 | None |
[9]:
dataset_list = [
'tgs00101', # life expectancy
'tgs00026', # exposable income
'tgs00036', # primary income
'tgs00010', # unemployment rate
'tgs00112', # touristic bed number
]
Download and aggregate data
To make the subsequent analysis easier, we download all data at once and store it in a single dataframe. Additionally, we store associated meta data in another dataframe.
[11]:
df_list = []
meta_list = []
for dataset in tqdm(dataset_list):
# get dataset description
dataset_name = eurostat.label_eurostat_tables(dataset)[0]
print(f'Parsing {dataset} ({dataset_name})')
# retrieve data
df_data = eurostat.get_eurostat(dataset, time_format='raw')
df_data = eurostat.label_eurostat(df_data, code=['geo'], fix_duplicated=True)
# index rows
common_columns = ['geo_code', 'geo']
df_data['idx'] = df_data[common_columns].apply(
lambda row: '_'.join(row.values.astype(str)), axis=1
)
# identify meta information
info_columns = (
set(df_data.columns)
- set(common_columns)
- {'idx', 'time', 'values', 'info', 'info_short'}
)
meta_cols = [
';'.join(
f'{k}={row._asdict()[k]}'
for k in sorted(row._asdict().keys())
if k != 'Index'
)
for row in df_data[info_columns].itertuples()
]
meta_idx, meta_label = pd.factorize(meta_cols)
df_data['meta'] = [
f'{dataset}_{time}_{x}' for x, time in zip(meta_idx, df_data['time'])
]
# pivot data
df_piv = df_data.pivot(index='idx', columns='meta', values='values')
# store data
df_list.append(df_piv)
meta_list.extend(
[
{
'dataset': dataset,
'name': dataset_name,
'meta_idx': idx,
'meta_label': label,
}
for idx, label in zip(np.unique(meta_idx), meta_label)
]
)
Parsing tgs00101 (Life expectancy at birth by sex and NUTS 2 region)
Parsing tgs00026 (Disposable income of private households by NUTS 2 regions)
Parsing tgs00036 (Primary income of private households by NUTS 2 regions)
Parsing tgs00010 (Unemployment rate by NUTS 2 regions)
Parsing tgs00112 (Number of establishments and bed-places by NUTS 2 regions)
Each row corresponds to a geographic region, and each column to a certain statistic.
[13]:
df_all = pd.concat(df_list, axis=1)
df_all.head()
[13]:
| meta | tgs00101_2007_0 | tgs00101_2007_1 | tgs00101_2007_2 | tgs00101_2008_0 | tgs00101_2008_1 | tgs00101_2008_2 | tgs00101_2009_0 | tgs00101_2009_1 | tgs00101_2009_2 | tgs00101_2010_0 | ... | tgs00112_2015_0 | tgs00112_2015_1 | tgs00112_2016_0 | tgs00112_2016_1 | tgs00112_2017_0 | tgs00112_2017_1 | tgs00112_2018_0 | tgs00112_2018_1 | tgs00112_2019_0 | tgs00112_2019_1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| idx | |||||||||||||||||||||
| AT11_Burgenland (AT) | 83.3 | 76.2 | 79.7 | 83.4 | 76.7 | 80.0 | 84.2 | 77.5 | 80.9 | 83.9 | ... | 28414.0 | 436.0 | 28252.0 | 427.0 | 28293.0 | 427.0 | 27551.0 | 418.0 | 27404.0 | 412.0 |
| AT12_Niederösterreich | 82.7 | 77.0 | 79.9 | 82.8 | 77.1 | 80.0 | 82.7 | 77.4 | 80.1 | 83.3 | ... | 68399.0 | 1459.0 | 68714.0 | 1429.0 | 67465.0 | 1434.0 | 67868.0 | 1435.0 | 68246.0 | 1447.0 |
| AT13_Wien | 82.0 | 76.7 | 79.5 | 82.4 | 77.0 | 79.9 | 82.1 | 76.5 | 79.4 | 82.2 | ... | 73901.0 | 604.0 | 74771.0 | 1072.0 | 75462.0 | 1212.0 | 78672.0 | 1735.0 | 79245.0 | 2070.0 |
| AT21_Kärnten | 83.6 | 77.8 | 80.8 | 84.1 | 77.7 | 81.0 | 83.7 | 77.7 | 80.8 | 84.0 | ... | 142508.0 | 2765.0 | 141191.0 | 2695.0 | 140473.0 | 2717.0 | 140448.0 | 2720.0 | 140165.0 | 2716.0 |
| AT22_Steiermark | 83.6 | 77.6 | 80.7 | 83.7 | 77.5 | 80.6 | 83.5 | 77.6 | 80.6 | 83.9 | ... | 112499.0 | 2432.0 | 116262.0 | 2471.0 | 118592.0 | 2471.0 | 125850.0 | 2481.0 | 128815.0 | 2521.0 |
5 rows × 120 columns
Extra information for each column is stored in an additional dataframe.
[14]:
df_meta = pd.DataFrame(meta_list)
df_meta.head()
[14]:
| dataset | name | meta_idx | meta_label | |
|---|---|---|---|---|
| 0 | tgs00101 | Life expectancy at birth by sex and NUTS 2 region | 0 | age=Less than 1 year;sex=Females;unit=Year |
| 1 | tgs00101 | Life expectancy at birth by sex and NUTS 2 region | 1 | age=Less than 1 year;sex=Males;unit=Year |
| 2 | tgs00101 | Life expectancy at birth by sex and NUTS 2 region | 2 | age=Less than 1 year;sex=Total;unit=Year |
| 3 | tgs00026 | Disposable income of private households by NUT... | 0 | direct=Balance;na_item=Disposable income, net;... |
| 4 | tgs00036 | Primary income of private households by NUTS 2... | 0 | direct=Balance;na_item=Balance of primary inco... |
[15]:
def get_meta(col_name, extended=False):
dataset, time, idx = col_name.split('_')
res = df_meta[(df_meta['dataset'] == dataset) & (df_meta['meta_idx'] == int(idx))]
assert res.shape[0] == 1
res = res.iloc[0]
if extended:
return f'{res["name"]}\n({res["meta_label"]})'
else:
return res["name"]
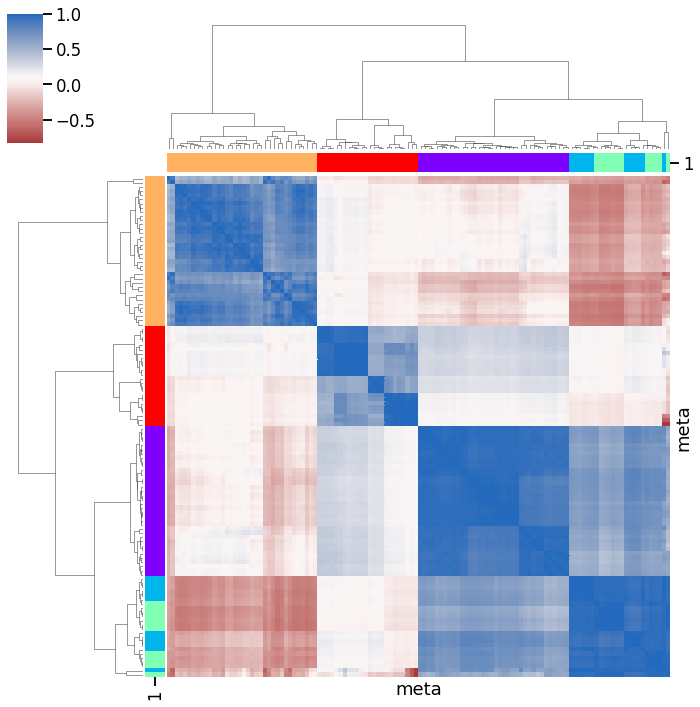
Overview
To get a rough overview of the data, we can plot the clustered correlation matrix.
[16]:
df_corr = df_all.corr()
[17]:
non_nan_datasets = set(df_corr.dropna(axis=0).index) & set(
df_corr.dropna(axis=1).columns
)
df_corr = df_corr.loc[non_nan_datasets, non_nan_datasets]
[18]:
# assign unique color to each data source
cluster_names = df_all.columns.str.split('_').str[0].unique()
cluser_colors = cm.rainbow(np.linspace(0, 1, len(cluster_names))).tolist()
cluster_map = {n: c for n, c in zip(cluster_names, cluser_colors)}
cluster_colors = pd.DataFrame(
[(col, cluster_map[col.split('_')[0]]) for col in df_all.columns]
).set_index(0)[1]
[19]:
g = sns.clustermap(
df_corr,
xticklabels=False,
yticklabels=False,
cmap='vlag_r',
row_colors=cluster_colors,
col_colors=cluster_colors,
)
Data preparation
In order to combine both geographical and statistical data, we merge the data…
[20]:
df_all['geo_code'] = [idx.split('_')[0] for idx in df_all.index]
[21]:
df_merged = df_geo.merge(df_all, left_on='NUTS_ID', right_on='geo_code')
df_merged.head()
[21]:
| id | CNTR_CODE | NUTS_NAME | LEVL_CODE | FID | NUTS_ID | geometry | geo | tgs00101_2007_0 | tgs00101_2007_1 | ... | tgs00112_2015_1 | tgs00112_2016_0 | tgs00112_2016_1 | tgs00112_2017_0 | tgs00112_2017_1 | tgs00112_2018_0 | tgs00112_2018_1 | tgs00112_2019_0 | tgs00112_2019_1 | geo_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | DE24 | DE | Oberfranken | 2 | DE24 | DE24 | POLYGON ((11.48157 50.43162, 11.48290 50.40161... | DE24 | 82.2 | 76.7 | ... | 890.0 | 40555.0 | 876.0 | 40804.0 | 869.0 | 40049.0 | 863.0 | 41574.0 | 865.0 | DE24 |
| 1 | NO03 | NO | Sør-Østlandet | 2 | NO03 | NO03 | POLYGON ((10.60072 60.13161, 10.57604 60.11572... | NO03 | 82.3 | 77.7 | ... | 464.0 | 143032.0 | 455.0 | 142724.0 | 421.0 | 151654.0 | 413.0 | NaN | NaN | NO03 |
| 2 | HU11 | HU | Budapest | 2 | HU11 | HU11 | POLYGON ((18.93274 47.57117, 19.02713 47.58717... | HU11 | 78.7 | 72.4 | ... | 342.0 | 57210.0 | 359.0 | NaN | NaN | 56213.0 | 375.0 | 58711.0 | 379.0 | HU11 |
| 3 | HU12 | HU | Pest | 2 | HU12 | HU12 | POLYGON ((18.96591 47.02896, 18.89943 47.07825... | HU12 | 77.9 | 69.8 | ... | 229.0 | 16348.0 | 232.0 | NaN | NaN | 17816.0 | 248.0 | 16927.0 | 240.0 | HU12 |
| 4 | HU21 | HU | Közép-Dunántúl | 2 | HU21 | HU21 | POLYGON ((18.68843 47.57707, 18.70448 47.52134... | HU21 | 77.8 | 69.4 | ... | 610.0 | 63641.0 | 625.0 | 65846.0 | 651.0 | 70627.0 | 685.0 | 69919.0 | 692.0 | HU21 |
5 rows × 129 columns
…and dissolve it to reach a resolution which leads to a nice visualization.
[22]:
%%time
df_diss = df_merged.dissolve(by='CNTR_CODE', aggfunc='mean')
CPU times: user 612 ms, sys: 17.3 ms, total: 630 ms
Wall time: 663 ms
[23]:
df_diss.head()
[23]:
| geometry | LEVL_CODE | tgs00101_2007_0 | tgs00101_2007_1 | tgs00101_2007_2 | tgs00101_2008_0 | tgs00101_2008_1 | tgs00101_2008_2 | tgs00101_2009_0 | tgs00101_2009_1 | ... | tgs00112_2015_0 | tgs00112_2015_1 | tgs00112_2016_0 | tgs00112_2016_1 | tgs00112_2017_0 | tgs00112_2017_1 | tgs00112_2018_0 | tgs00112_2018_1 | tgs00112_2019_0 | tgs00112_2019_1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNTR_CODE | |||||||||||||||||||||
| AT | POLYGON ((15.99624 46.83540, 15.98885 46.82563... | 2 | 83.366667 | 77.611111 | 80.566667 | 83.622222 | 77.977778 | 80.866667 | 83.600000 | 77.844444 | ... | 110385.000000 | 2257.222222 | 111271.333333 | 2291.000000 | 112577.666667 | 2320.555556 | 116181.888889 | 2388.222222 | 115356.444444 | 2439.000000 |
| BE | POLYGON ((6.02490 50.18278, 5.97197 50.17189, ... | 2 | 82.581818 | 76.909091 | 79.790909 | 82.536364 | 76.627273 | 79.600000 | 82.672727 | 77.190909 | ... | 33499.272727 | 726.363636 | 33497.090909 | 746.363636 | 33982.000000 | 779.090909 | 35438.727273 | 837.363636 | 35962.272727 | 877.363636 |
| BG | POLYGON ((26.56154 41.92627, 26.58121 41.90292... | 2 | 76.466667 | 69.400000 | 72.833333 | 76.850000 | 69.683333 | 73.133333 | 77.183333 | 70.000000 | ... | 53744.166667 | 533.666667 | 54710.666667 | 555.166667 | 58120.666667 | 557.666667 | 55932.833333 | 576.333333 | 56917.666667 | 610.666667 |
| CH | POLYGON ((9.15938 46.16960, 9.12055 46.13610, ... | 2 | 84.557143 | 79.614286 | 82.185714 | 84.785714 | 79.942857 | 82.471429 | 84.742857 | 80.014286 | ... | NaN | NaN | 97329.571429 | 5902.714286 | NaN | NaN | 94951.857143 | 5579.571429 | NaN | NaN |
| CY | POLYGON ((33.75237 34.97711, 33.70164 34.97605... | 2 | 82.100000 | 77.600000 | 79.800000 | 82.900000 | 78.200000 | 80.600000 | 83.500000 | 78.500000 | ... | 85414.000000 | 788.000000 | 84239.000000 | 785.000000 | 85965.000000 | 796.000000 | 87240.000000 | 802.000000 | 90188.000000 | 816.000000 |
5 rows × 122 columns
Visualization
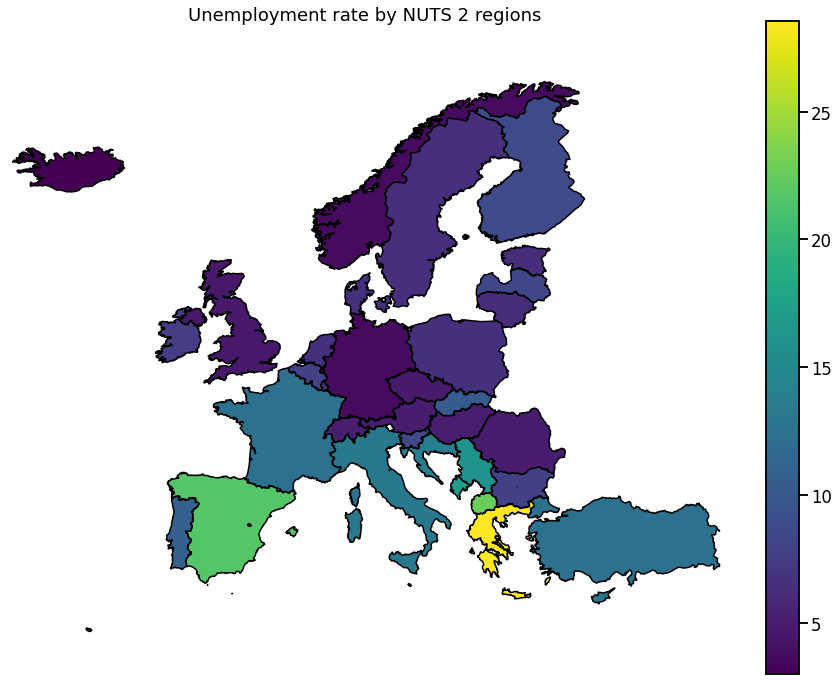
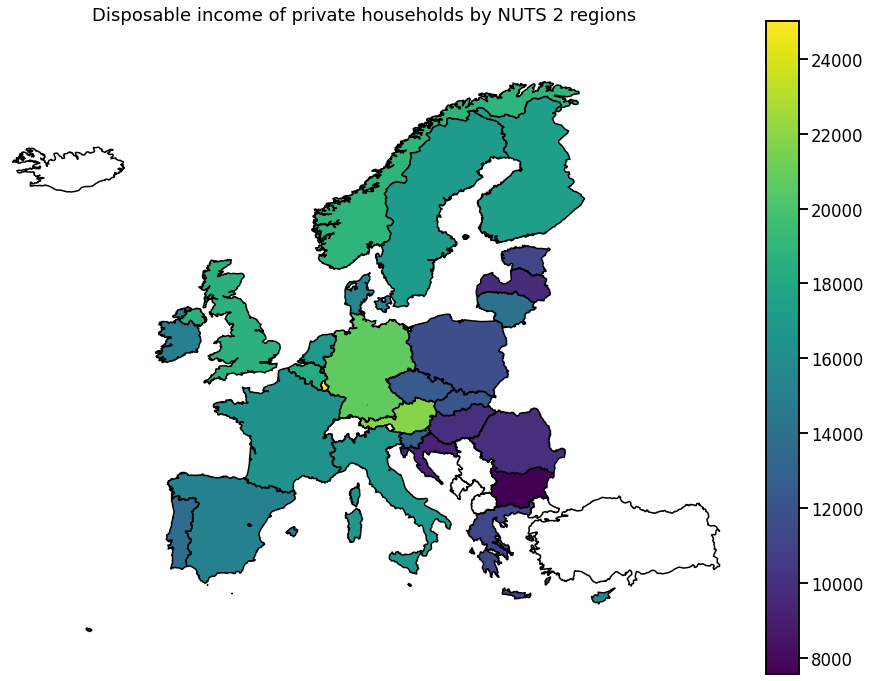
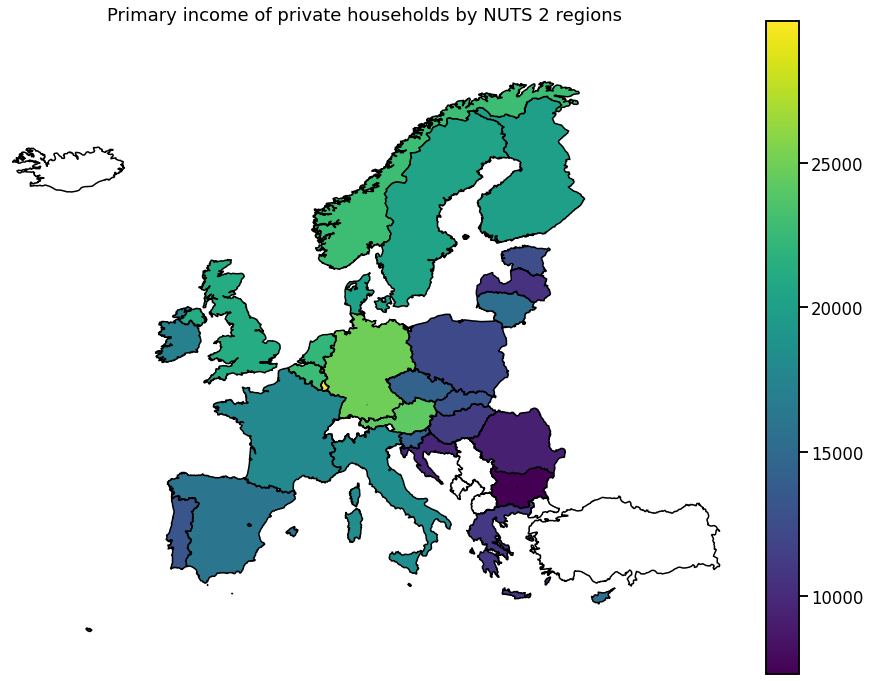
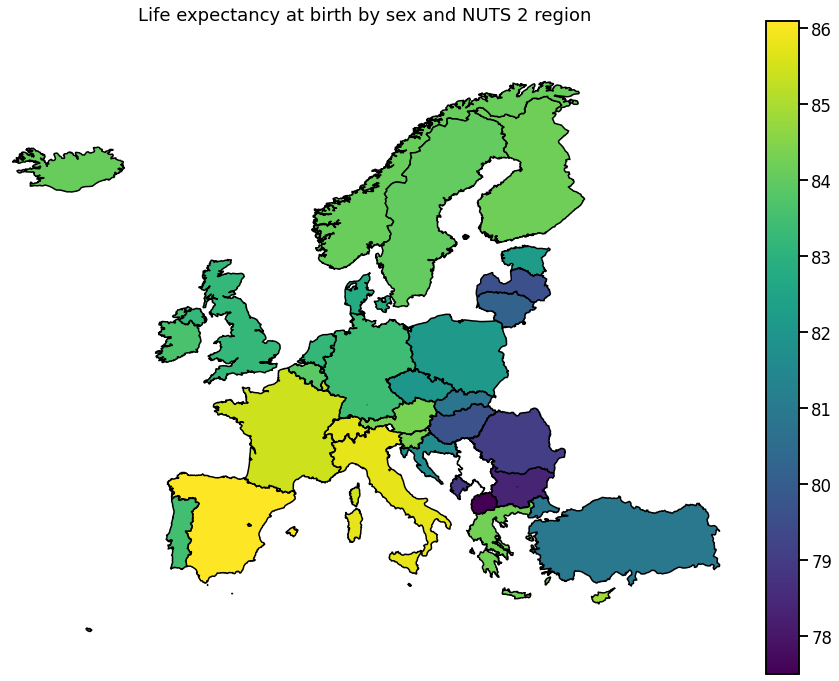
We visualize the data using choropleth maps. By doing so, each geographical region is colored according to a statistic of interest.
In the following, we will use a single entry from each downloaded dataset.
[24]:
time = 2016
hue_selection = [
f'{row.dataset}_{time}_{row.meta_idx}'
for row in df_meta.groupby('dataset').apply(lambda x: x.sample(n=1)).itertuples()
]
hue_selection
[24]:
['tgs00010_2016_0',
'tgs00026_2016_0',
'tgs00036_2016_0',
'tgs00101_2016_0',
'tgs00112_2016_0']
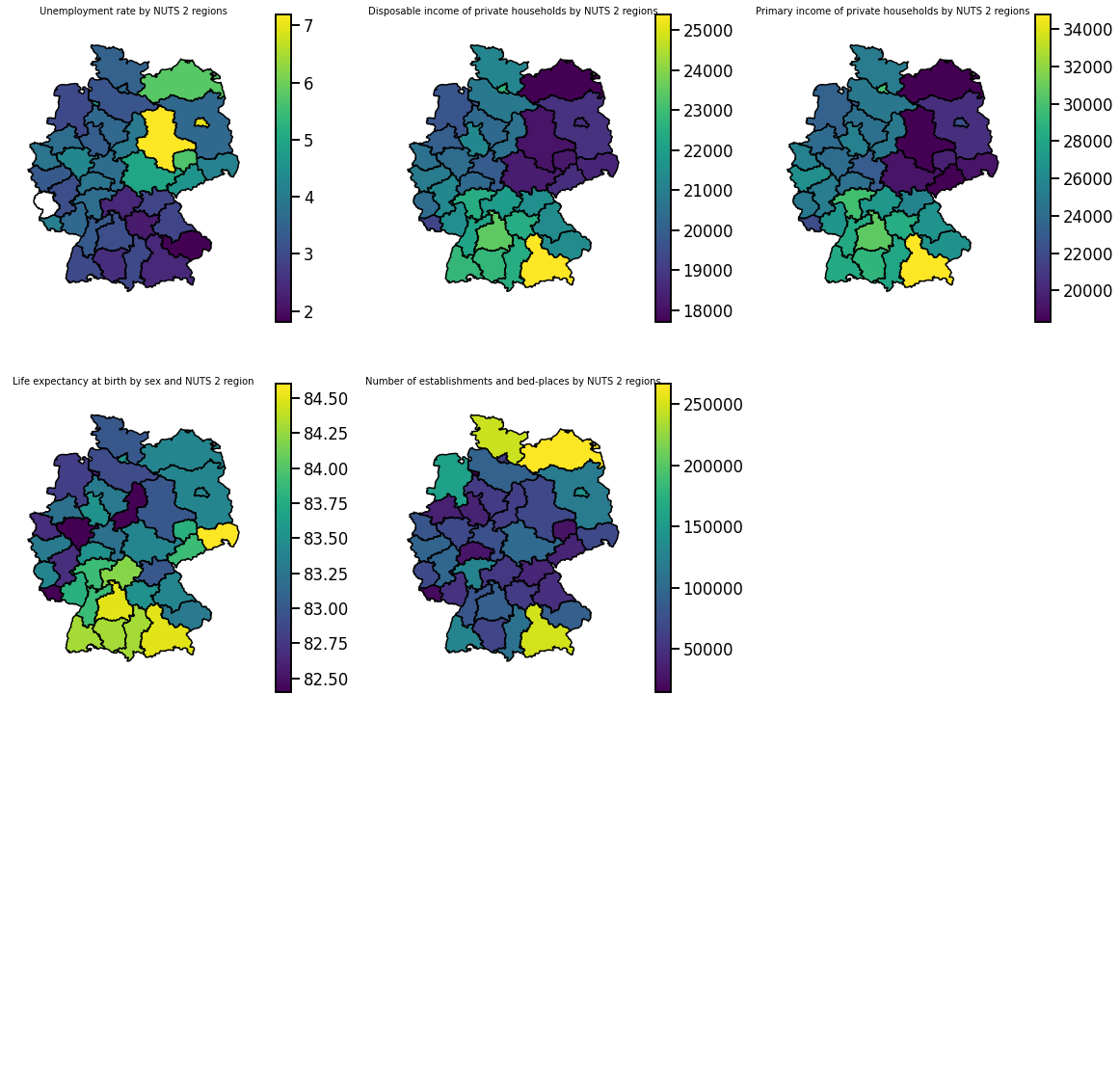
Single country
We can get a precise overview of individual regions within a single country.
[25]:
size = int(np.ceil(np.sqrt(len(hue_selection)))) # size of plot grid
fig, axes = plt.subplots(nrows=size, ncols=size, figsize=(20, 20))
[ax.axis('off') for ax in axes.ravel()]
for hue, ax in zip(hue_selection, axes.ravel()):
sub = df_merged[(df_merged['CNTR_CODE'] == 'DE')]
gplt.choropleth(sub, hue=hue, legend=True, ax=ax)
ax.set_aspect(1.4)
ax.set_title(get_meta(hue), fontsize=10)
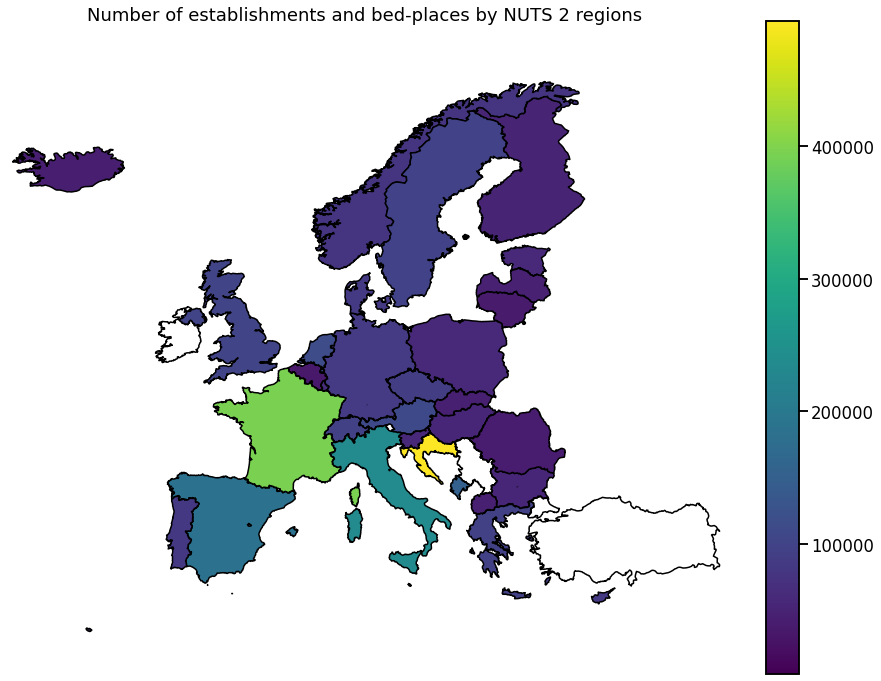
Europe
To get a more global picture, we can also plot the aggregated data per country.
[26]:
for hue in hue_selection:
ax = gplt.choropleth(
df_diss,
hue=hue,
legend=True, # k=5,
figsize=(16, 12),
extent=(
-25,
30,
45,
75,
), # (min_longitude, min_latitude, max_longitude, max_latitude)
)
ax.set_aspect(1.4)
ax.set_title(get_meta(hue))