Comparing threading and multiprocessing
[1]:
import time
import multiprocessing as mp
from multiprocessing import Pool as ProcessPool
from multiprocessing.pool import ThreadPool
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
[2]:
sns.set_context('talk')
Introduction
There are various choices when trying to run code in parallel.
The threading module will run all threads on the same CPU core which requires less overhead and allows for more efficient sharing of memory. However, it is not truly parallel and executes the threads when others are idling.
The multiprocessing module runs the processes on multiple CPU cores and can thus execute code at the same time.
Preparations
To investigate the differences between threading and multiprocessing, we will simulate work for each data point and measure when it was executed.
Due to some design choices, we have to import the worker function from a separate module for the ProcessPool to work.
[3]:
%%writefile worker.py
import time
def worker(data):
tmp = []
for i in data:
# simulate CPU load
for _ in range(1_000_000):
pass
# store execution time
tmp.append(time.time())
return tmp
Overwriting worker.py
[4]:
from worker import worker
[5]:
data = list(range(20))
num = mp.cpu_count()
Computations
We run the worker function on the dataset for each executor in both the process and thread pool.
[6]:
%%time
with ThreadPool(num) as p:
thread_result = p.map(worker, [data] * num)
CPU times: user 3.79 s, sys: 131 ms, total: 3.92 s
Wall time: 3.86 s
[7]:
%%time
with ProcessPool(num) as p:
process_result = p.map(worker, [data] * num)
CPU times: user 26 ms, sys: 56.7 ms, total: 82.7 ms
Wall time: 1.4 s
Next, we store the result in a dataframe.
[8]:
df_thread = pd.melt(
pd.DataFrame(thread_result, index=[f'job {i:2}' for i in range(num)]).T
)
df_thread['type'] = 'thread'
df_process = pd.melt(
pd.DataFrame(process_result, index=[f'job {i:2}' for i in range(num)]).T
)
df_process['type'] = 'process'
df = pd.concat([df_thread, df_process], ignore_index=True)
df['value'] = df.groupby('type')['value'].apply(lambda x: x - x.min()) # normalize time
df.head()
[8]:
| variable | value | type | |
|---|---|---|---|
| 0 | job 0 | 0.105189 | thread |
| 1 | job 0 | 0.316431 | thread |
| 2 | job 0 | 0.592710 | thread |
| 3 | job 0 | 0.704985 | thread |
| 4 | job 0 | 0.822085 | thread |
Investigation
There are two main observations:
multiprocessingexecution takes less total runtime thanthreadingmultiprocessingtimestamps are in parallel, whilethreadingtimestamps are serial
[9]:
g = sns.FacetGrid(df, row='type', aspect=2)
g.map_dataframe(sns.scatterplot, x='value', y='variable')
g.set_axis_labels('Time [s]', 'Pool')
[9]:
<seaborn.axisgrid.FacetGrid at 0x140feb100>
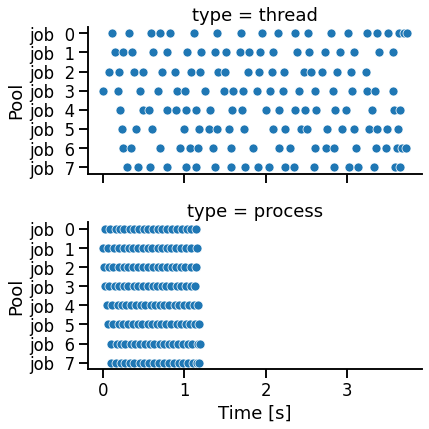
The decision whether to use threading or multiprocessing depends on the use case.
As a general rule of thumb, one should use threading if the problem is IO bound and multiprocessing if it is CPU bound.